If you want to get help directly related to a page/query, use the Info Pages / Medium Info Pages. Direct links are available at each (Query) Page.
If you are completely lost, here is link to a short description of what CSB.DB is and is not.
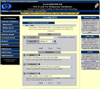
Enter this page.
Values based filter options 
If you select a specific sub-class of the class 'Value' you retrieve all genes associated by co-response, which meet pre-set thresholds. This thresholds are applied to the degree to which the genes are related and / or to statistic parameters. Default settings are given.
positive & p.value < 0.01 & power > 0.7: This values based sub-class can only be selected if you chosen one of the following correlation coefficients: Spearman, Pearson or Kendall. You retrieve all genes positively associated by co-response with a probability of at least 99% (p.value < 0.01) and a power of test of at least 70% (power > 0.7).
positive & best 10%: This values based sub-class is available for all types of coefficients. You retrieve all genes which are in the range of the 10% strongest positive co-responses. For correlation coefficients values near +1 will be retrieved. Distance near 0 will be retrieved for distance coefficients of into distance range converted coefficients.
positive & p.value < 0.05: This values based sub-class can only be selected if you chosen one of the following correlation coefficients: Spearman, Pearson or Kendall. You retrieve all genes positively associated by co-response with a probability of at least 95% (p.value < 0.05).
negative & p.value < 0.01 & power > 0.7: This values based sub-class can only be selected if you chosen one of the following correlation coefficients: Spearman, Pearson or Kendall. You retrieve all genes negatively associated by co-response with a probability of at least 99% (p.value < 0.01) and a power of test of at least 70% (power > 0.7).
negative & best 10%: This values based sub-class can only be seleted if yo chosen one of the correlation coefficients. You retrieve all genes which are in the range of the 10% strongest negative co-responses. Values near -1 will be retrieved.
negative & p.value < 0.05: This values based sub-class can only be selected if you chosen one of the following correlation coefficients: Spearman, Pearson or Kendall. You retrieve all genes negatively associated by co-response with a probability of at least 95% (p.value < 0.05).
Category based filter options
If you select a specific sub-class of the class 'Category' (if available) you retrieve all genes associated by co-response, which are assigned to this functional category. The functional categories are self-explanatory. Below the table you can found a graph of the normalised distribution of co-responses belonging to the individual categories.




